lstsq(a, b, rcond='warn')
Computes the vector x that approximately solves the equation a @ x = b
. The equation may be under-, well-, or over-determined (i.e., the number of linearly independent rows of a can be less than, equal to, or greater than its number of linearly independent columns). If a is square and of full rank, then x (but for round-off error) is the "exact" solution of the equation. Else, x minimizes the Euclidean 2-norm $||b - ax||$
. If there are multiple minimizing solutions, the one with the smallest 2-norm $||x||$
is returned.
If b is a matrix, then all array results are returned as matrices.
"Coefficient" matrix.
Ordinate or "dependent variable" values. If b is two-dimensional, the least-squares solution is calculated for each of the K
columns of b.
Cut-off ratio for small singular values of a. For the purposes of rank determination, singular values are treated as zero if they are smaller than :None:None:`rcond` times the largest singular value of a.
If not set, a FutureWarning is given. The previous default of -1
will use the machine precision as :None:None:`rcond` parameter, the new default will use the machine precision times :None:None:`max(M, N)`. To silence the warning and use the new default, use rcond=None
, to keep using the old behavior, use rcond=-1
.
If computation does not converge.
Least-squares solution. If b is two-dimensional, the solutions are in the K
columns of x.
Sums of squared residuals: Squared Euclidean 2-norm for each column in b - a @ x
. If the rank of a is < N or M <= N, this is an empty array. If b is 1-dimensional, this is a (1,) shape array. Otherwise the shape is (K,).
Rank of matrix a.
Singular values of a.
Return the least-squares solution to a linear matrix equation.
scipy.linalg.lstsq
Similar function in SciPy.
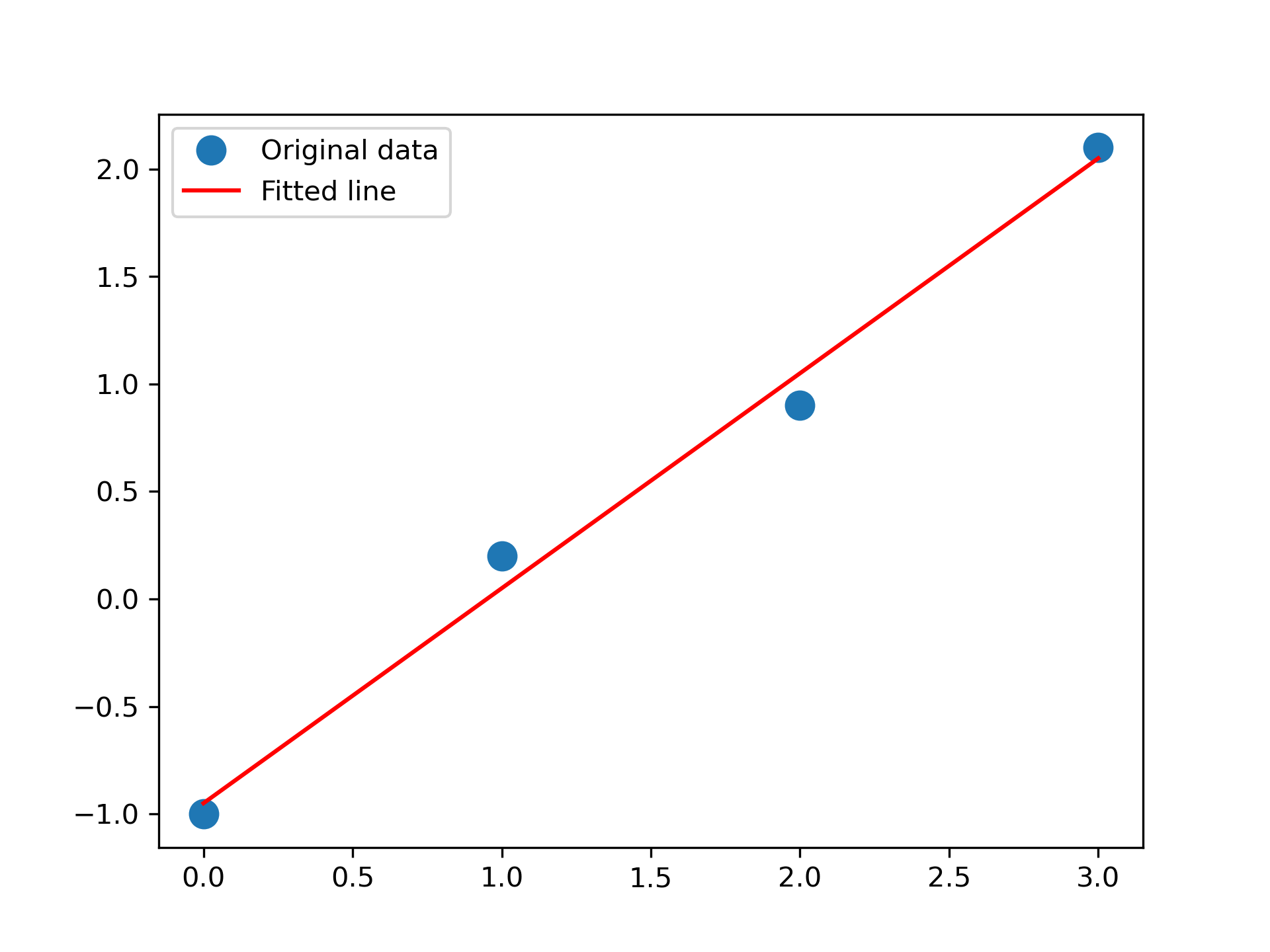
Fit a line, y = mx + c
, through some noisy data-points:
>>> x = np.array([0, 1, 2, 3])
... y = np.array([-1, 0.2, 0.9, 2.1])
By examining the coefficients, we see that the line should have a gradient of roughly 1 and cut the y-axis at, more or less, -1.
We can rewrite the line equation as y = Ap
, where A = [[x 1]]
and p = [[m], [c]]
. Now use lstsq
to solve for :None:None:`p`:
>>> A = np.vstack([x, np.ones(len(x))]).T
... A array([[ 0., 1.], [ 1., 1.], [ 2., 1.], [ 3., 1.]])
>>> m, c = np.linalg.lstsq(A, y, rcond=None)[0]
... m, c (1.0 -0.95) # may vary
Plot the data along with the fitted line:
>>> import matplotlib.pyplot as plt
... _ = plt.plot(x, y, 'o', label='Original data', markersize=10)
... _ = plt.plot(x, m*x + c, 'r', label='Fitted line')
... _ = plt.legend()
... plt.show()
The following pages refer to to this document either explicitly or contain code examples using this.
numpy.polynomial.polynomial.polyfit
numpy.ma.extras.polyfit
numpy.polynomial.hermite_e.hermefit
numpy.linalg.qr
numpy.polyfit
numpy.polynomial.chebyshev.chebfit
numpy.linalg.solve
numpy.polynomial.hermite.hermfit
numpy.polynomial.legendre.legfit
numpy.polynomial.laguerre.lagfit
numpy.linalg.lstsq
Hover to see nodes names; edges to Self not shown, Caped at 50 nodes.
Using a canvas is more power efficient and can get hundred of nodes ; but does not allow hyperlinks; , arrows or text (beyond on hover)
SVG is more flexible but power hungry; and does not scale well to 50 + nodes.
All aboves nodes referred to, (or are referred from) current nodes; Edges from Self to other have been omitted (or all nodes would be connected to the central node "self" which is not useful). Nodes are colored by the library they belong to, and scaled with the number of references pointing them