logspace(start, stop, num=50, endpoint=True, base=10.0, dtype=None, axis=0)
In linear space, the sequence starts at base ** start
(:None:None:`base` to the power of :None:None:`start`) and ends with base ** stop
(see :None:None:`endpoint` below).
Non-scalar :None:None:`start` and :None:None:`stop` are now supported.
Logspace is equivalent to the code
>>> y = np.linspace(start, stop, num=num, endpoint=endpoint) ... # doctest: +SKIP >>> power(base, y).astype(dtype) ... # doctest: +SKIP
base ** start
is the starting value of the sequence.
base ** stop
is the final value of the sequence, unless :None:None:`endpoint` is False. In that case, num + 1
values are spaced over the interval in log-space, of which all but the last (a sequence of length :None:None:`num`) are returned.
Number of samples to generate. Default is 50.
If true, :None:None:`stop` is the last sample. Otherwise, it is not included. Default is True.
The base of the log space. The step size between the elements in ln(samples) / ln(base)
(or log_base(samples)
) is uniform. Default is 10.0.
The type of the output array. If dtype
is not given, the data type is inferred from :None:None:`start` and :None:None:`stop`. The inferred type will never be an integer; :None:None:`float` is chosen even if the arguments would produce an array of integers.
The axis in the result to store the samples. Relevant only if start or stop are array-like. By default (0), the samples will be along a new axis inserted at the beginning. Use -1 to get an axis at the end.
:None:None:`num` samples, equally spaced on a log scale.
Return numbers spaced evenly on a log scale.
arange
Similar to linspace, with the step size specified instead of the number of samples. Note that, when used with a float endpoint, the endpoint may or may not be included.
geomspace
Similar to logspace, but with endpoints specified directly.
linspace
Similar to logspace, but with the samples uniformly distributed in linear space, instead of log space.
>>> np.logspace(2.0, 3.0, num=4) array([ 100. , 215.443469 , 464.15888336, 1000. ])
>>> np.logspace(2.0, 3.0, num=4, endpoint=False) array([100. , 177.827941 , 316.22776602, 562.34132519])
>>> np.logspace(2.0, 3.0, num=4, base=2.0) array([4. , 5.0396842 , 6.34960421, 8. ])
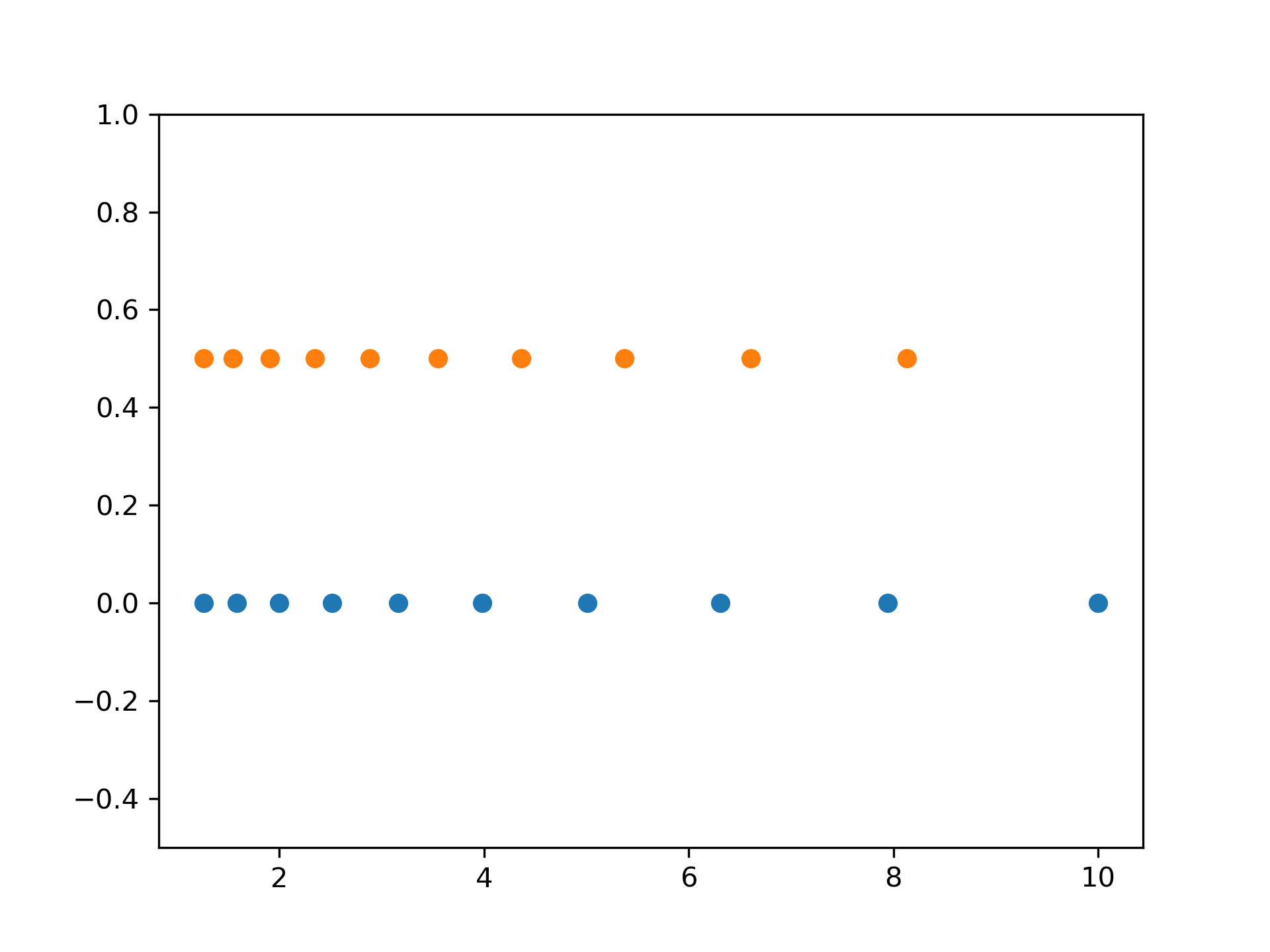
Graphical illustration:
>>> import matplotlib.pyplot as plt
... N = 10
... x1 = np.logspace(0.1, 1, N, endpoint=True)
... x2 = np.logspace(0.1, 1, N, endpoint=False)
... y = np.zeros(N)
... plt.plot(x1, y, 'o') [<matplotlib.lines.Line2D object at 0x...>]
>>> plt.plot(x2, y + 0.5, 'o') [<matplotlib.lines.Line2D object at 0x...>]
>>> plt.ylim([-0.5, 1]) (-0.5, 1)
>>> plt.show()
The following pages refer to to this document either explicitly or contain code examples using this.
scipy.signal._filter_design.freqs
numpy.geomspace
numpy.linspace
scipy.signal._filter_design.ellipord
numpy.lib.scimath.log
scipy.signal._filter_design.buttord
scipy.signal._filter_design.freqs_zpk
Hover to see nodes names; edges to Self not shown, Caped at 50 nodes.
Using a canvas is more power efficient and can get hundred of nodes ; but does not allow hyperlinks; , arrows or text (beyond on hover)
SVG is more flexible but power hungry; and does not scale well to 50 + nodes.
All aboves nodes referred to, (or are referred from) current nodes; Edges from Self to other have been omitted (or all nodes would be connected to the central node "self" which is not useful). Nodes are colored by the library they belong to, and scaled with the number of references pointing them