csd(x, y, fs=1.0, window='hann', nperseg=None, noverlap=None, nfft=None, detrend='constant', return_onesided=True, scaling='density', axis=-1, average='mean')
By convention, Pxy is computed with the conjugate FFT of X multiplied by the FFT of Y.
If the input series differ in length, the shorter series will be zero-padded to match.
An appropriate amount of overlap will depend on the choice of window and on your requirements. For the default Hann window an overlap of 50% is a reasonable trade off between accurately estimating the signal power, while not over counting any of the data. Narrower windows may require a larger overlap.
Time series of measurement values
Time series of measurement values
Sampling frequency of the x and y time series. Defaults to 1.0.
Desired window to use. If :None:None:`window` is a string or tuple, it is passed to get_window
to generate the window values, which are DFT-even by default. See get_window
for a list of windows and required parameters. If :None:None:`window` is array_like it will be used directly as the window and its length must be nperseg. Defaults to a Hann window.
Length of each segment. Defaults to None, but if window is str or tuple, is set to 256, and if window is array_like, is set to the length of the window.
Number of points to overlap between segments. If :None:None:`None`, noverlap = nperseg // 2
. Defaults to :None:None:`None`.
Length of the FFT used, if a zero padded FFT is desired. If :None:None:`None`, the FFT length is :None:None:`nperseg`. Defaults to :None:None:`None`.
Specifies how to detrend each segment. If detrend
is a string, it is passed as the :None:None:`type` argument to the detrend
function. If it is a function, it takes a segment and returns a detrended segment. If detrend
is :None:None:`False`, no detrending is done. Defaults to 'constant'.
If :None:None:`True`, return a one-sided spectrum for real data. If :None:None:`False` return a two-sided spectrum. Defaults to :None:None:`True`, but for complex data, a two-sided spectrum is always returned.
Selects between computing the cross spectral density ('density') where :None:None:`Pxy` has units of V**2/Hz and computing the cross spectrum ('spectrum') where :None:None:`Pxy` has units of V**2, if x and y are measured in V and :None:None:`fs` is measured in Hz. Defaults to 'density'
Axis along which the CSD is computed for both inputs; the default is over the last axis (i.e. axis=-1
).
Method to use when averaging periodograms. If the spectrum is complex, the average is computed separately for the real and imaginary parts. Defaults to 'mean'.
Array of sample frequencies.
Cross spectral density or cross power spectrum of x,y.
Estimate the cross power spectral density, Pxy, using Welch's method.
coherence
Magnitude squared coherence by Welch's method.
lombscargle
Lomb-Scargle periodogram for unevenly sampled data
periodogram
Simple, optionally modified periodogram
welch
Power spectral density by Welch's method. [Equivalent to csd(x,x)]
>>> from scipy import signal
... import matplotlib.pyplot as plt
... rng = np.random.default_rng()
Generate two test signals with some common features.
>>> fs = 10e3
... N = 1e5
... amp = 20
... freq = 1234.0
... noise_power = 0.001 * fs / 2
... time = np.arange(N) / fs
... b, a = signal.butter(2, 0.25, 'low')
... x = rng.normal(scale=np.sqrt(noise_power), size=time.shape)
... y = signal.lfilter(b, a, x)
... x += amp*np.sin(2*np.pi*freq*time)
... y += rng.normal(scale=0.1*np.sqrt(noise_power), size=time.shape)
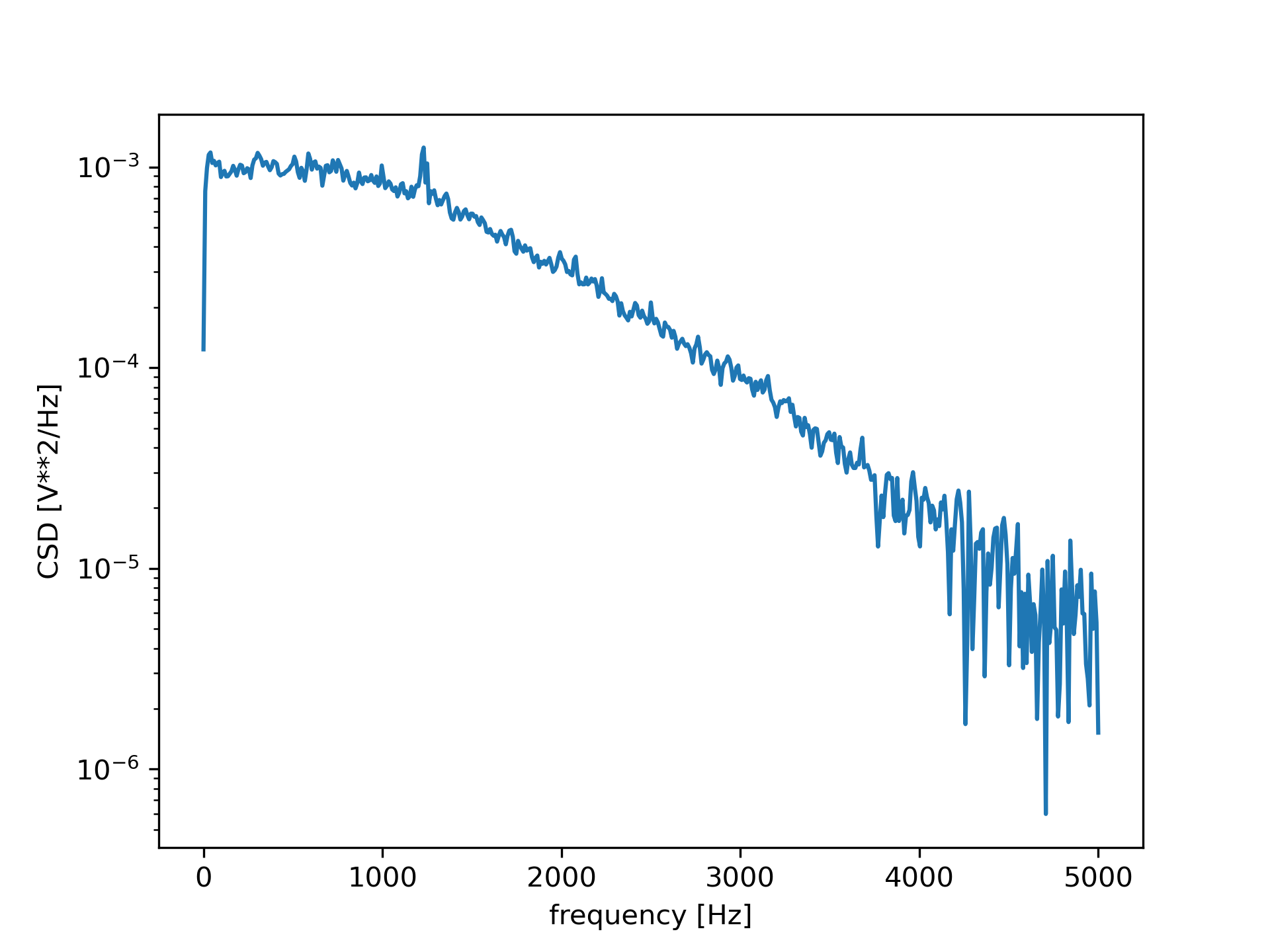
Compute and plot the magnitude of the cross spectral density.
>>> f, Pxy = signal.csd(x, y, fs, nperseg=1024)
... plt.semilogy(f, np.abs(Pxy))
... plt.xlabel('frequency [Hz]')
... plt.ylabel('CSD [V**2/Hz]')
... plt.show()
The following pages refer to to this document either explicitly or contain code examples using this.
scipy.signal._spectral_py.coherence
scipy.signal._spectral_py.stft
scipy.signal._spectral_py.csd
scipy.signal._spectral_py.spectrogram
scipy.signal._spectral_py.lombscargle
Hover to see nodes names; edges to Self not shown, Caped at 50 nodes.
Using a canvas is more power efficient and can get hundred of nodes ; but does not allow hyperlinks; , arrows or text (beyond on hover)
SVG is more flexible but power hungry; and does not scale well to 50 + nodes.
All aboves nodes referred to, (or are referred from) current nodes; Edges from Self to other have been omitted (or all nodes would be connected to the central node "self" which is not useful). Nodes are colored by the library they belong to, and scaled with the number of references pointing them