For more information, see the :None:None:`FITPACK_` site about this function.
<Unimplemented 'target' '.. _FITPACK: http://www.netlib.org/dierckx/sphere.f'>
1-D sequences of data points (order is not important). Coordinates must be given in radians. Theta must lie within the interval [0, pi]
, and phi must lie within the interval [0, 2pi]
.
Positive 1-D sequence of weights.
Positive smoothing factor defined for estimation condition: sum((w(i)*(r(i) - s(theta(i), phi(i))))**2, axis=0) <= s
Default s=len(w)
which should be a good value if 1/w[i]
is an estimate of the standard deviation of r[i]
.
A threshold for determining the effective rank of an over-determined linear system of equations. :None:None:`eps` should have a value within the open interval (0, 1)
, the default is 1e-16.
Smooth bivariate spline approximation in spherical coordinates.
BivariateSpline
a base class for bivariate splines.
LSQBivariateSpline
a bivariate spline using weighted least-squares fitting
LSQSphereBivariateSpline
a bivariate spline in spherical coordinates using weighted least-squares fitting
RectBivariateSpline
a bivariate spline over a rectangular mesh.
RectSphereBivariateSpline
a bivariate spline over a rectangular mesh on a sphere
SmoothBivariateSpline
a smoothing bivariate spline through the given points
UnivariateSpline
a smooth univariate spline to fit a given set of data points.
bisplev
a function to evaluate a bivariate B-spline and its derivatives
bisplrep
a function to find a bivariate B-spline representation of a surface
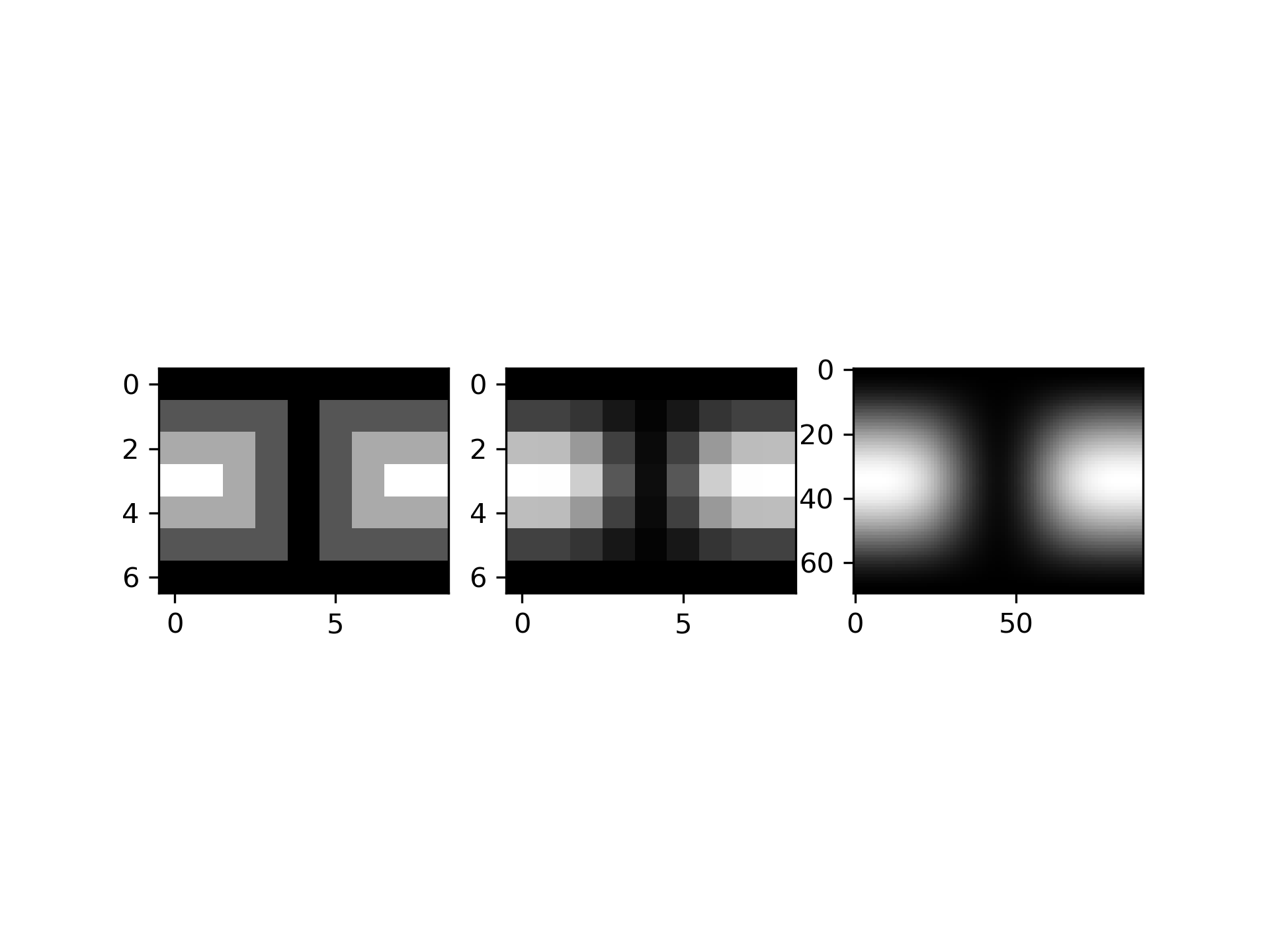
Suppose we have global data on a coarse grid (the input data does not have to be on a grid):
>>> theta = np.linspace(0., np.pi, 7)
... phi = np.linspace(0., 2*np.pi, 9)
... data = np.empty((theta.shape[0], phi.shape[0]))
... data[:,0], data[0,:], data[-1,:] = 0., 0., 0.
... data[1:-1,1], data[1:-1,-1] = 1., 1.
... data[1,1:-1], data[-2,1:-1] = 1., 1.
... data[2:-2,2], data[2:-2,-2] = 2., 2.
... data[2,2:-2], data[-3,2:-2] = 2., 2.
... data[3,3:-2] = 3.
... data = np.roll(data, 4, 1)
We need to set up the interpolator object
>>> lats, lons = np.meshgrid(theta, phi)
... from scipy.interpolate import SmoothSphereBivariateSpline
... lut = SmoothSphereBivariateSpline(lats.ravel(), lons.ravel(),
... data.T.ravel(), s=3.5)
As a first test, we'll see what the algorithm returns when run on the input coordinates
>>> data_orig = lut(theta, phi)
Finally we interpolate the data to a finer grid
>>> fine_lats = np.linspace(0., np.pi, 70)
... fine_lons = np.linspace(0., 2 * np.pi, 90)
>>> data_smth = lut(fine_lats, fine_lons)
>>> import matplotlib.pyplot as plt
... fig = plt.figure()
... ax1 = fig.add_subplot(131)
... ax1.imshow(data, interpolation='nearest')
... ax2 = fig.add_subplot(132)
... ax2.imshow(data_orig, interpolation='nearest')
... ax3 = fig.add_subplot(133)
... ax3.imshow(data_smth, interpolation='nearest')
... plt.show()
The following pages refer to to this document either explicitly or contain code examples using this.
scipy.interpolate._fitpack2.RectSphereBivariateSpline
scipy.interpolate._fitpack2.SmoothBivariateSpline
scipy.interpolate._fitpack2.BivariateSpline
scipy.interpolate._fitpack2.LSQSphereBivariateSpline
scipy.interpolate._fitpack2.RectBivariateSpline
scipy.interpolate._fitpack2.LSQBivariateSpline
scipy.interpolate._fitpack2.SmoothSphereBivariateSpline
scipy.interpolate._fitpack2.UnivariateSpline
Hover to see nodes names; edges to Self not shown, Caped at 50 nodes.
Using a canvas is more power efficient and can get hundred of nodes ; but does not allow hyperlinks; , arrows or text (beyond on hover)
SVG is more flexible but power hungry; and does not scale well to 50 + nodes.
All aboves nodes referred to, (or are referred from) current nodes; Edges from Self to other have been omitted (or all nodes would be connected to the central node "self" which is not useful). Nodes are colored by the library they belong to, and scaled with the number of references pointing them