odeint(func, y0, t, args=(), Dfun=None, col_deriv=0, full_output=0, ml=None, mu=None, rtol=None, atol=None, tcrit=None, h0=0.0, hmax=0.0, hmin=0.0, ixpr=0, mxstep=0, mxhnil=0, mxordn=12, mxords=5, printmessg=0, tfirst=False)
differential equation.
Solve a system of ordinary differential equations using lsoda from the FORTRAN library odepack.
Solves the initial value problem for stiff or non-stiff systems of first order ode-s:
dy/dt = func(y, t, ...) [or func(t, y, ...)]
where y can be a vector.
:None:None:`func` are in the opposite order of the arguments in the system definition function used by the :None:None:`scipy.integrate.ode` class and the function :None:None:`scipy.integrate.solve_ivp`. To use a function with the signature func(t, y, ...)
, the argument :None:None:`tfirst` must be set to True
.
If either of these are not None or non-negative, then the Jacobian is assumed to be banded. These give the number of lower and upper non-zero diagonals in this banded matrix. For the banded case, :None:None:`Dfun` should return a matrix whose rows contain the non-zero bands (starting with the lowest diagonal). Thus, the return matrix :None:None:`jac` from :None:None:`Dfun` should have shape (ml + mu + 1, len(y0))
when ml >=0
or mu >=0
. The data in :None:None:`jac` must be stored such that jac[i - j + mu, j]
holds the derivative of the :None:None:`i'th equation with respect to the 'j'th
state variable. If 'col_deriv` is True, the transpose of this :None:None:`jac` must be returned.
The input parameters :None:None:`rtol` and atol
determine the error control performed by the solver. The solver will control the vector, e, of estimated local errors in y, according to an inequality of the form max-norm of (e / ewt) <= 1
, where ewt is a vector of positive error weights computed as ewt = rtol * abs(y) + atol
. rtol and atol can be either vectors the same length as y or scalars. Defaults to 1.49012e-8.
Vector of critical points (e.g., singularities) where integration care should be taken.
The step size to be attempted on the first step.
The maximum absolute step size allowed.
The minimum absolute step size allowed.
Whether to generate extra printing at method switches.
Maximum number of (internally defined) steps allowed for each integration point in t.
Maximum number of messages printed.
Maximum order to be allowed for the non-stiff (Adams) method.
Maximum order to be allowed for the stiff (BDF) method.
Computes the derivative of y at t. If the signature is callable(t, y, ...)
, then the argument :None:None:`tfirst` must be set True
.
Initial condition on y (can be a vector).
A sequence of time points for which to solve for y. The initial value point should be the first element of this sequence. This sequence must be monotonically increasing or monotonically decreasing; repeated values are allowed.
Extra arguments to pass to function.
Gradient (Jacobian) of :None:None:`func`. If the signature is callable(t, y, ...)
, then the argument :None:None:`tfirst` must be set True
.
True if :None:None:`Dfun` defines derivatives down columns (faster), otherwise :None:None:`Dfun` should define derivatives across rows.
True if to return a dictionary of optional outputs as the second output
Whether to print the convergence message
If True, the first two arguments of :None:None:`func` (and :None:None:`Dfun`, if given) must t, y
instead of the default y, t
.
Array containing the value of y for each desired time in t, with the initial value y0
in the first row.
Dictionary containing additional output information
======= ============================================================ key meaning ======= ============================================================ 'hu' vector of step sizes successfully used for each time step 'tcur' vector with the value of t reached for each time step (will always be at least as large as the input times) 'tolsf' vector of tolerance scale factors, greater than 1.0, computed when a request for too much accuracy was detected 'tsw' value of t at the time of the last method switch (given for each time step) 'nst' cumulative number of time steps 'nfe' cumulative number of function evaluations for each time step 'nje' cumulative number of jacobian evaluations for each time step 'nqu' a vector of method orders for each successful step 'imxer' index of the component of largest magnitude in the weighted local error vector (e / ewt) on an error return, -1 otherwise 'lenrw' the length of the double work array required 'leniw' the length of integer work array required 'mused' a vector of method indicators for each successful time step: 1: adams (nonstiff), 2: bdf (stiff) ======= ============================================================
Integrate a system of ordinary differential equations.
ode
a more object-oriented integrator based on VODE
quad
for finding the area under a curve
solve_ivp
solve an initial value problem for a system of ODEs
theta''(t) + b*theta'(t) + c*sin(theta(t)) = 0
theta'(t) = omega(t) omega'(t) = -b*omega(t) - c*sin(theta(t))
Let y be the vector [`theta`, omega
]. We implement this system in Python as:
>>> def pend(y, t, b, c):
... theta, omega = y
... dydt = [omega, -b*omega - c*np.sin(theta)]
... return dydt ...
We assume the constants are b = 0.25 and c = 5.0:
>>> b = 0.25
... c = 5.0
For initial conditions, we assume the pendulum is nearly vertical with :None:None:`theta(0)` = :None:None:`pi` - 0.1, and is initially at rest, so :None:None:`omega(0)` = 0. Then the vector of initial conditions is
>>> y0 = [np.pi - 0.1, 0.0]
We will generate a solution at 101 evenly spaced samples in the interval 0 <= t <= 10. So our array of times is:
>>> t = np.linspace(0, 10, 101)
Call odeint
to generate the solution. To pass the parameters b and c to pend
, we give them to odeint
using the :None:None:`args` argument.
>>> from scipy.integrate import odeint
... sol = odeint(pend, y0, t, args=(b, c))
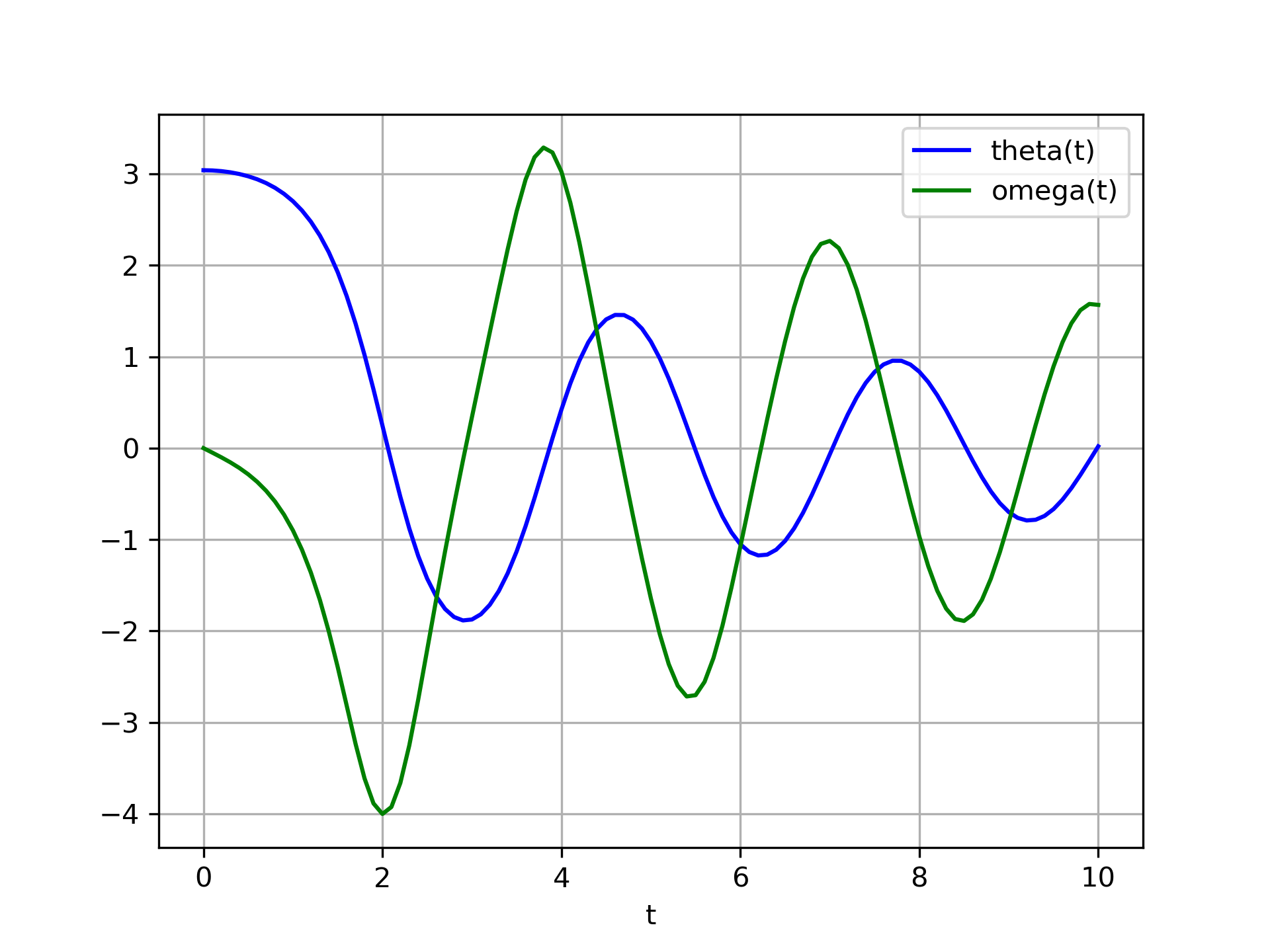
The solution is an array with shape (101, 2). The first column is :None:None:`theta(t)`, and the second is :None:None:`omega(t)`. The following code plots both components.
>>> import matplotlib.pyplot as plt
... plt.plot(t, sol[:, 0], 'b', label='theta(t)')
... plt.plot(t, sol[:, 1], 'g', label='omega(t)')
... plt.legend(loc='best')
... plt.xlabel('t')
... plt.grid()
... plt.show()
The following pages refer to to this document either explicitly or contain code examples using this.
scipy.signal._ltisys.lsim2
scipy.integrate._odepack_py.odeint
scipy.signal._ltisys.step2
scipy.integrate._ode.ode
scipy.signal._ltisys.impulse2
Hover to see nodes names; edges to Self not shown, Caped at 50 nodes.
Using a canvas is more power efficient and can get hundred of nodes ; but does not allow hyperlinks; , arrows or text (beyond on hover)
SVG is more flexible but power hungry; and does not scale well to 50 + nodes.
All aboves nodes referred to, (or are referred from) current nodes; Edges from Self to other have been omitted (or all nodes would be connected to the central node "self" which is not useful). Nodes are colored by the library they belong to, and scaled with the number of references pointing them