solve_ivp(fun, t_span, y0, method='RK45', t_eval=None, dense_output=False, events=None, vectorized=False, args=None, **options)
This function numerically integrates a system of ordinary differential equations given an initial value:
dy / dt = f(t, y) y(t0) = y0
Here t is a 1-D independent variable (time), y(t) is an N-D vector-valued function (state), and an N-D vector-valued function f(t, y) determines the differential equations. The goal is to find y(t) approximately satisfying the differential equations, given an initial value y(t0)=y0.
Some of the solvers support integration in the complex domain, but note that for stiff ODE solvers, the right-hand side must be complex-differentiable (satisfy Cauchy-Riemann equations ). To solve a problem in the complex domain, pass y0 with a complex data type. Another option always available is to rewrite your problem for real and imaginary parts separately.
Right-hand side of the system. The calling signature is fun(t, y)
. Here t is a scalar, and there are two options for the ndarray y: It can either have shape (n,); then :None:None:`fun` must return array_like with shape (n,). Alternatively, it can have shape (n, k); then :None:None:`fun` must return an array_like with shape (n, k), i.e., each column corresponds to a single column in y. The choice between the two options is determined by :None:None:`vectorized` argument (see below). The vectorized implementation allows a faster approximation of the Jacobian by finite differences (required for stiff solvers).
Interval of integration (t0, tf). The solver starts with t=t0 and integrates until it reaches t=tf.
Initial state. For problems in the complex domain, pass y0
with a complex data type (even if the initial value is purely real).
Integration method to use:
'RK45' (default): Explicit Runge-Kutta method of order 5(4) . The error is controlled assuming accuracy of the fourth-order method, but steps are taken using the fifth-order accurate formula (local extrapolation is done). A quartic interpolation polynomial is used for the dense output . Can be applied in the complex domain.
'RK23': Explicit Runge-Kutta method of order 3(2) . The error is controlled assuming accuracy of the second-order method, but steps are taken using the third-order accurate formula (local extrapolation is done). A cubic Hermite polynomial is used for the dense output. Can be applied in the complex domain.
'DOP853': Explicit Runge-Kutta method of order 8 . Python implementation of the "DOP853" algorithm originally written in Fortran . A 7-th order interpolation polynomial accurate to 7-th order is used for the dense output. Can be applied in the complex domain.
'Radau': Implicit Runge-Kutta method of the Radau IIA family of order 5 . The error is controlled with a third-order accurate embedded formula. A cubic polynomial which satisfies the collocation conditions is used for the dense output.
'BDF': Implicit multi-step variable-order (1 to 5) method based on a backward differentiation formula for the derivative approximation . The implementation follows the one described in . A quasi-constant step scheme is used and accuracy is enhanced using the NDF modification. Can be applied in the complex domain.
'LSODA': Adams/BDF method with automatic stiffness detection and switching , . This is a wrapper of the Fortran solver from ODEPACK.
Explicit Runge-Kutta methods ('RK23', 'RK45', 'DOP853') should be used for non-stiff problems and implicit methods ('Radau', 'BDF') for stiff problems . Among Runge-Kutta methods, 'DOP853' is recommended for solving with high precision (low values of :None:None:`rtol` and atol
).
If not sure, first try to run 'RK45'. If it makes unusually many iterations, diverges, or fails, your problem is likely to be stiff and you should use 'Radau' or 'BDF'. 'LSODA' can also be a good universal choice, but it might be somewhat less convenient to work with as it wraps old Fortran code.
You can also pass an arbitrary class derived from OdeSolver
which implements the solver.
Times at which to store the computed solution, must be sorted and lie within :None:None:`t_span`. If None (default), use points selected by the solver.
Whether to compute a continuous solution. Default is False.
Events to track. If None (default), no events will be tracked. Each event occurs at the zeros of a continuous function of time and state. Each function must have the signature event(t, y)
and return a float. The solver will find an accurate value of t at which event(t, y(t)) = 0
using a root-finding algorithm. By default, all zeros will be found. The solver looks for a sign change over each step, so if multiple zero crossings occur within one step, events may be missed. Additionally each :None:None:`event` function might have the following attributes:
terminal: bool, optional
Whether to terminate integration if this event occurs. Implicitly False if not assigned.
direction: float, optional
Direction of a zero crossing. If
:None:None:`direction`is positive,:None:None:`event`will only trigger when going from negative to positive, and vice versa if:None:None:`direction`is negative. If 0, then either direction will trigger event. Implicitly 0 if not assigned.
You can assign attributes like event.terminal = True
to any function in Python.
Whether :None:None:`fun` is implemented in a vectorized fashion. Default is False.
Additional arguments to pass to the user-defined functions. If given, the additional arguments are passed to all user-defined functions. So if, for example, :None:None:`fun` has the signature fun(t, y, a, b, c)
, then :None:None:`jac` (if given) and any event functions must have the same signature, and :None:None:`args` must be a tuple of length 3.
Options passed to a chosen solver. All options available for already implemented solvers are listed below.
Initial step size. Default is :None:None:`None` which means that the algorithm should choose.
Maximum allowed step size. Default is np.inf, i.e., the step size is not bounded and determined solely by the solver.
Relative and absolute tolerances. The solver keeps the local error estimates less than atol + rtol * abs(y)
. Here :None:None:`rtol` controls a relative accuracy (number of correct digits), while atol
controls absolute accuracy (number of correct decimal places). To achieve the desired :None:None:`rtol`, set atol
to be lower than the lowest value that can be expected from rtol * abs(y)
so that :None:None:`rtol` dominates the allowable error. If atol
is larger than rtol * abs(y)
the number of correct digits is not guaranteed. Conversely, to achieve the desired atol
set :None:None:`rtol` such that rtol * abs(y)
is always lower than atol
. If components of y have different scales, it might be beneficial to set different atol
values for different components by passing array_like with shape (n,) for atol
. Default values are 1e-3 for :None:None:`rtol` and 1e-6 for atol
.
Jacobian matrix of the right-hand side of the system with respect to y, required by the 'Radau', 'BDF' and 'LSODA' method. The Jacobian matrix has shape (n, n) and its element (i, j) is equal to d f_i / d y_j
. There are three ways to define the Jacobian:
If array_like or sparse_matrix, the Jacobian is assumed to be constant. Not supported by 'LSODA'.
If callable, the Jacobian is assumed to depend on both t and y; it will be called as
jac(t, y), as necessary. For 'Radau' and 'BDF' methods, the return value might be a sparse matrix.If None (default), the Jacobian will be approximated by finite differences.
It is generally recommended to provide the Jacobian rather than relying on a finite-difference approximation.
Defines a sparsity structure of the Jacobian matrix for a finite- difference approximation. Its shape must be (n, n). This argument is ignored if :None:None:`jac` is not :None:None:`None`. If the Jacobian has only few non-zero elements in each row, providing the sparsity structure will greatly speed up the computations . A zero entry means that a corresponding element in the Jacobian is always zero. If None (default), the Jacobian is assumed to be dense. Not supported by 'LSODA', see :None:None:`lband` and :None:None:`uband` instead.
Parameters defining the bandwidth of the Jacobian for the 'LSODA' method, i.e., jac[i, j] != 0 only for i - lband <= j <= i + uband
. Default is None. Setting these requires your jac routine to return the Jacobian in the packed format: the returned array must have n
columns and uband + lband + 1
rows in which Jacobian diagonals are written. Specifically jac_packed[uband + i - j , j] = jac[i, j]
. The same format is used in scipy.linalg.solve_banded
(check for an illustration). These parameters can be also used with jac=None
to reduce the number of Jacobian elements estimated by finite differences.
The minimum allowed step size for 'LSODA' method. By default :None:None:`min_step` is zero.
Time points.
Values of the solution at t.
Found solution as OdeSolution
instance; None if dense_output
was set to False.
Contains for each event type a list of arrays at which an event of that type event was detected. None if :None:None:`events` was None.
For each value of :None:None:`t_events`, the corresponding value of the solution. None if :None:None:`events` was None.
Number of evaluations of the right-hand side.
Number of evaluations of the Jacobian.
Number of LU decompositions.
Reason for algorithm termination:
-1: Integration step failed.
0: The solver successfully reached the end of
:None:None:`tspan`.1: A termination event occurred.
Human-readable description of the termination reason.
True if the solver reached the interval end or a termination event occurred ( status >= 0
).
Solve an initial value problem for a system of ODEs.
Basic exponential decay showing automatically chosen time points.
>>> from scipy.integrate import solve_ivp
... def exponential_decay(t, y): return -0.5 * y
... sol = solve_ivp(exponential_decay, [0, 10], [2, 4, 8])
... print(sol.t) [ 0. 0.11487653 1.26364188 3.06061781 4.81611105 6.57445806 8.33328988 10. ]
>>> print(sol.y) [[2. 1.88836035 1.06327177 0.43319312 0.18017253 0.07483045 0.03107158 0.01350781] [4. 3.7767207 2.12654355 0.86638624 0.36034507 0.14966091 0.06214316 0.02701561] [8. 7.5534414 4.25308709 1.73277247 0.72069014 0.29932181 0.12428631 0.05403123]]
Specifying points where the solution is desired.
>>> sol = solve_ivp(exponential_decay, [0, 10], [2, 4, 8],
... t_eval=[0, 1, 2, 4, 10])
... print(sol.t) [ 0 1 2 4 10]
>>> print(sol.y) [[2. 1.21305369 0.73534021 0.27066736 0.01350938] [4. 2.42610739 1.47068043 0.54133472 0.02701876] [8. 4.85221478 2.94136085 1.08266944 0.05403753]]
Cannon fired upward with terminal event upon impact. The terminal
and direction
fields of an event are applied by monkey patching a function. Here y[0]
is position and y[1]
is velocity. The projectile starts at position 0 with velocity +10. Note that the integration never reaches t=100 because the event is terminal.
>>> def upward_cannon(t, y): return [y[1], -0.5]
... def hit_ground(t, y): return y[0]
... hit_ground.terminal = True
... hit_ground.direction = -1
... sol = solve_ivp(upward_cannon, [0, 100], [0, 10], events=hit_ground)
... print(sol.t_events) [array([40.])]
>>> print(sol.t) [0.00000000e+00 9.99900010e-05 1.09989001e-03 1.10988901e-02 1.11088891e-01 1.11098890e+00 1.11099890e+01 4.00000000e+01]
Use dense_output
and :None:None:`events` to find position, which is 100, at the apex of the cannonball's trajectory. Apex is not defined as terminal, so both apex and hit_ground are found. There is no information at t=20, so the sol attribute is used to evaluate the solution. The sol attribute is returned by setting dense_output=True
. Alternatively, the :None:None:`y_events` attribute can be used to access the solution at the time of the event.
>>> def apex(t, y): return y[1]
... sol = solve_ivp(upward_cannon, [0, 100], [0, 10],
... events=(hit_ground, apex), dense_output=True)
... print(sol.t_events) [array([40.]), array([20.])]
>>> print(sol.t) [0.00000000e+00 9.99900010e-05 1.09989001e-03 1.10988901e-02 1.11088891e-01 1.11098890e+00 1.11099890e+01 4.00000000e+01]
>>> print(sol.sol(sol.t_events[1][0])) [100. 0.]
>>> print(sol.y_events) [array([[-5.68434189e-14, -1.00000000e+01]]), array([[1.00000000e+02, 1.77635684e-15]])]
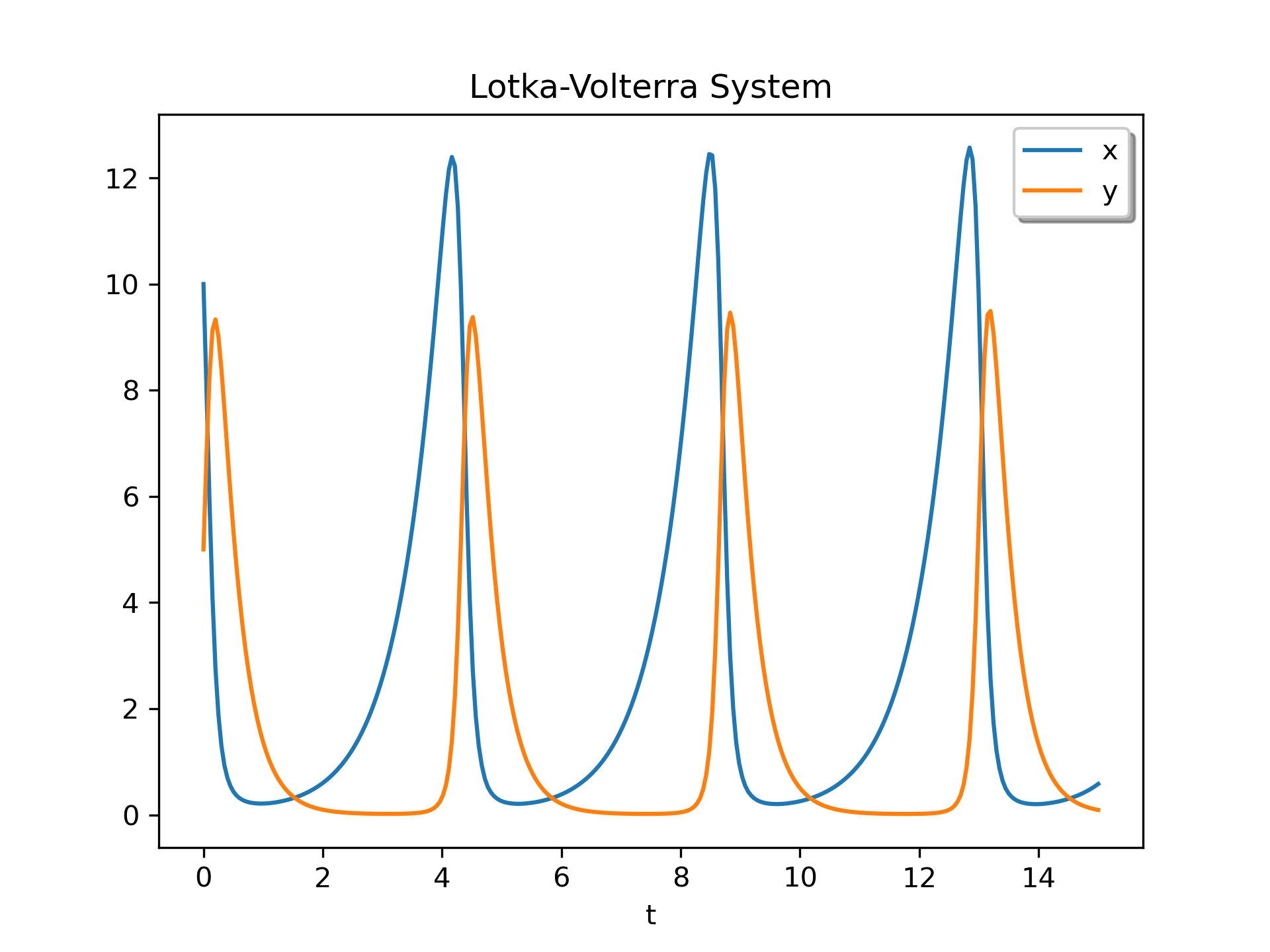
As an example of a system with additional parameters, we'll implement the Lotka-Volterra equations .
>>> def lotkavolterra(t, z, a, b, c, d):
... x, y = z
... return [a*x - b*x*y, -c*y + d*x*y] ...
We pass in the parameter values a=1.5, b=1, c=3 and d=1 with the :None:None:`args` argument.
>>> sol = solve_ivp(lotkavolterra, [0, 15], [10, 5], args=(1.5, 1, 3, 1),
... dense_output=True)
Compute a dense solution and plot it.
>>> t = np.linspace(0, 15, 300)
... z = sol.sol(t)
... import matplotlib.pyplot as plt
... plt.plot(t, z.T)
... plt.xlabel('t')
... plt.legend(['x', 'y'], shadow=True)
... plt.title('Lotka-Volterra System')
... plt.show()
The following pages refer to to this document either explicitly or contain code examples using this.
scipy.integrate._odepack_py.odeint
scipy.integrate._ivp.ivp.find_active_events
scipy.integrate._ivp.ivp.solve_ivp
Hover to see nodes names; edges to Self not shown, Caped at 50 nodes.
Using a canvas is more power efficient and can get hundred of nodes ; but does not allow hyperlinks; , arrows or text (beyond on hover)
SVG is more flexible but power hungry; and does not scale well to 50 + nodes.
All aboves nodes referred to, (or are referred from) current nodes; Edges from Self to other have been omitted (or all nodes would be connected to the central node "self" which is not useful). Nodes are colored by the library they belong to, and scaled with the number of references pointing them