>>> """
=======================
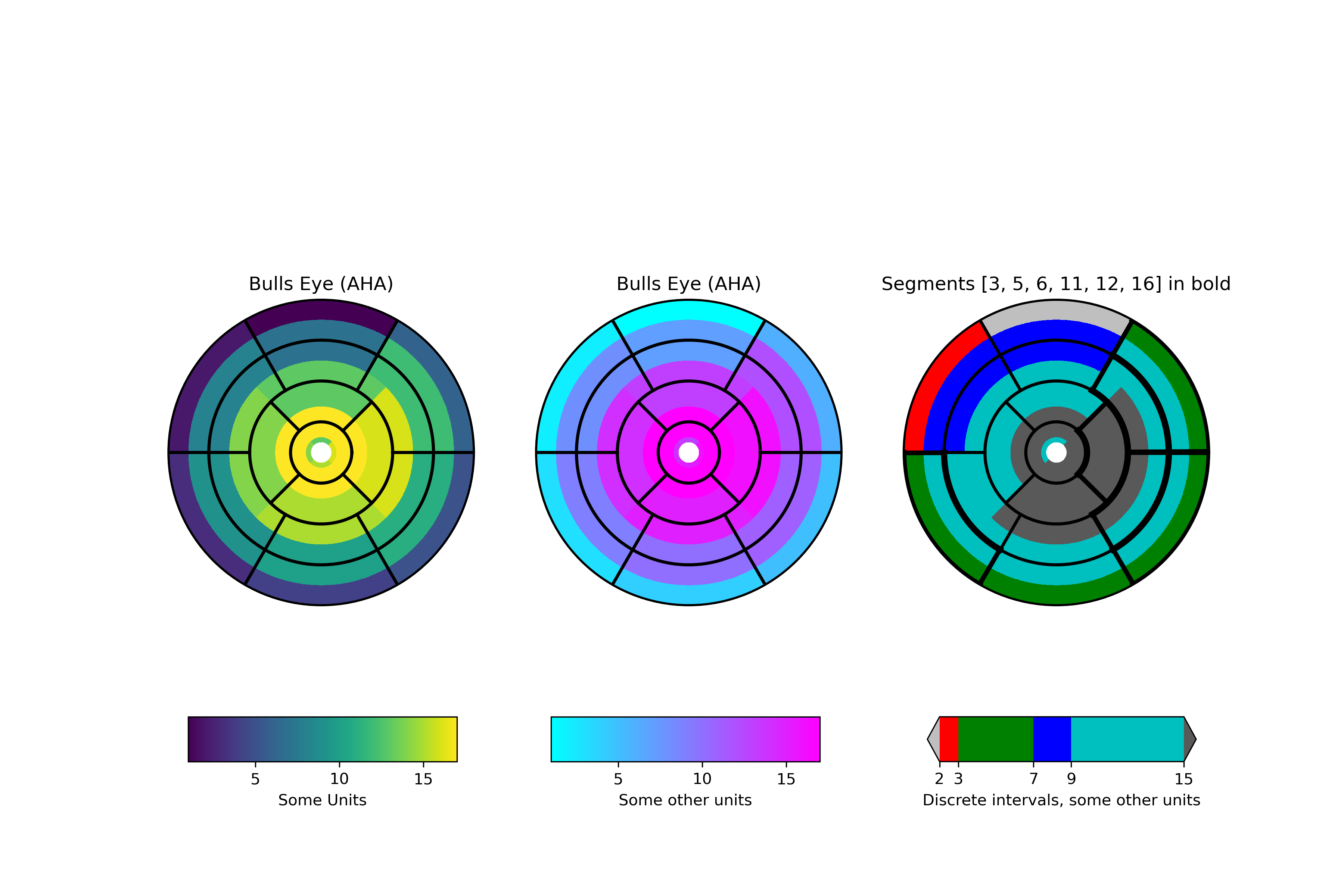
Left ventricle bullseye
=======================
This example demonstrates how to create the 17 segment model for the left
ventricle recommended by the American Heart Association (AHA).
"""
...
... import numpy as np
... import matplotlib as mpl
... import matplotlib.pyplot as plt
...
...
... def bullseye_plot(ax, data, seg_bold=None, cmap=None, norm=None):
... """
Bullseye representation for the left ventricle.
Parameters
----------
ax : axes
data : list of int and float
The intensity values for each of the 17 segments
seg_bold : list of int, optional
A list with the segments to highlight
cmap : ColorMap or None, optional
Optional argument to set the desired colormap
norm : Normalize or None, optional
Optional argument to normalize data into the [0.0, 1.0] range
Notes
-----
This function creates the 17 segment model for the left ventricle according
to the American Heart Association (AHA) [1]_
References
----------
.. [1] M. D. Cerqueira, N. J. Weissman, V. Dilsizian, A. K. Jacobs,
S. Kaul, W. K. Laskey, D. J. Pennell, J. A. Rumberger, T. Ryan,
and M. S. Verani, "Standardized myocardial segmentation and
nomenclature for tomographic imaging of the heart",
Circulation, vol. 105, no. 4, pp. 539-542, 2002.
"""
... if seg_bold is None:
... seg_bold = []
...
... linewidth = 2
... data = np.ravel(data)
...
... if cmap is None:
... cmap = plt.cm.viridis
...
... if norm is None:
... norm = mpl.colors.Normalize(vmin=data.min(), vmax=data.max())
...
... theta = np.linspace(0, 2 * np.pi, 768)
... r = np.linspace(0.2, 1, 4)
...
... # Create the bound for the segment 17
... for i in range(r.shape[0]):
... ax.plot(theta, np.repeat(r[i], theta.shape), '-k', lw=linewidth)
...
... # Create the bounds for the segments 1-12
... for i in range(6):
... theta_i = np.deg2rad(i * 60)
... ax.plot([theta_i, theta_i], [r[1], 1], '-k', lw=linewidth)
...
... # Create the bounds for the segments 13-16
... for i in range(4):
... theta_i = np.deg2rad(i * 90 - 45)
... ax.plot([theta_i, theta_i], [r[0], r[1]], '-k', lw=linewidth)
...
... # Fill the segments 1-6
... r0 = r[2:4]
... r0 = np.repeat(r0[:, np.newaxis], 128, axis=1).T
... for i in range(6):
... # First segment start at 60 degrees
... theta0 = theta[i * 128:i * 128 + 128] + np.deg2rad(60)
... theta0 = np.repeat(theta0[:, np.newaxis], 2, axis=1)
... z = np.ones((128, 2)) * data[i]
... ax.pcolormesh(theta0, r0, z, cmap=cmap, norm=norm, shading='auto')
... if i + 1 in seg_bold:
... ax.plot(theta0, r0, '-k', lw=linewidth + 2)
... ax.plot(theta0[0], [r[2], r[3]], '-k', lw=linewidth + 1)
... ax.plot(theta0[-1], [r[2], r[3]], '-k', lw=linewidth + 1)
...
... # Fill the segments 7-12
... r0 = r[1:3]
... r0 = np.repeat(r0[:, np.newaxis], 128, axis=1).T
... for i in range(6):
... # First segment start at 60 degrees
... theta0 = theta[i * 128:i * 128 + 128] + np.deg2rad(60)
... theta0 = np.repeat(theta0[:, np.newaxis], 2, axis=1)
... z = np.ones((128, 2)) * data[i + 6]
... ax.pcolormesh(theta0, r0, z, cmap=cmap, norm=norm, shading='auto')
... if i + 7 in seg_bold:
... ax.plot(theta0, r0, '-k', lw=linewidth + 2)
... ax.plot(theta0[0], [r[1], r[2]], '-k', lw=linewidth + 1)
... ax.plot(theta0[-1], [r[1], r[2]], '-k', lw=linewidth + 1)
...
... # Fill the segments 13-16
... r0 = r[0:2]
... r0 = np.repeat(r0[:, np.newaxis], 192, axis=1).T
... for i in range(4):
... # First segment start at 45 degrees
... theta0 = theta[i * 192:i * 192 + 192] + np.deg2rad(45)
... theta0 = np.repeat(theta0[:, np.newaxis], 2, axis=1)
... z = np.ones((192, 2)) * data[i + 12]
... ax.pcolormesh(theta0, r0, z, cmap=cmap, norm=norm, shading='auto')
... if i + 13 in seg_bold:
... ax.plot(theta0, r0, '-k', lw=linewidth + 2)
... ax.plot(theta0[0], [r[0], r[1]], '-k', lw=linewidth + 1)
... ax.plot(theta0[-1], [r[0], r[1]], '-k', lw=linewidth + 1)
...
... # Fill the segments 17
... if data.size == 17:
... r0 = np.array([0, r[0]])
... r0 = np.repeat(r0[:, np.newaxis], theta.size, axis=1).T
... theta0 = np.repeat(theta[:, np.newaxis], 2, axis=1)
... z = np.ones((theta.size, 2)) * data[16]
... ax.pcolormesh(theta0, r0, z, cmap=cmap, norm=norm, shading='auto')
... if 17 in seg_bold:
... ax.plot(theta0, r0, '-k', lw=linewidth + 2)
...
... ax.set_ylim([0, 1])
... ax.set_yticklabels([])
... ax.set_xticklabels([])
...
...
... # Create the fake data
... data = np.arange(17) + 1
...
...
... # Make a figure and axes with dimensions as desired.
... fig, ax = plt.subplots(figsize=(12, 8), nrows=1, ncols=3,
... subplot_kw=dict(projection='polar'))
... fig.canvas.manager.set_window_title('Left Ventricle Bulls Eyes (AHA)')
...
... # Create the axis for the colorbars
... axl = fig.add_axes([0.14, 0.15, 0.2, 0.05])
... axl2 = fig.add_axes([0.41, 0.15, 0.2, 0.05])
... axl3 = fig.add_axes([0.69, 0.15, 0.2, 0.05])
...
...
... # Set the colormap and norm to correspond to the data for which
... # the colorbar will be used.
... cmap = mpl.cm.viridis
... norm = mpl.colors.Normalize(vmin=1, vmax=17)
... # Create an empty ScalarMappable to set the colorbar's colormap and norm.
... # The following gives a basic continuous colorbar with ticks and labels.
... fig.colorbar(mpl.cm.ScalarMappable(cmap=cmap, norm=norm),
... cax=axl, orientation='horizontal', label='Some Units')
...
...
... # And again for the second colorbar.
... cmap2 = mpl.cm.cool
... norm2 = mpl.colors.Normalize(vmin=1, vmax=17)
... fig.colorbar(mpl.cm.ScalarMappable(cmap=cmap2, norm=norm2),
... cax=axl2, orientation='horizontal', label='Some other units')
...
...
... # The second example illustrates the use of a ListedColormap, a
... # BoundaryNorm, and extended ends to show the "over" and "under"
... # value colors.
... cmap3 = (mpl.colors.ListedColormap(['r', 'g', 'b', 'c'])
... .with_extremes(over='0.35', under='0.75'))
... # If a ListedColormap is used, the length of the bounds array must be
... # one greater than the length of the color list. The bounds must be
... # monotonically increasing.
... bounds = [2, 3, 7, 9, 15]
... norm3 = mpl.colors.BoundaryNorm(bounds, cmap3.N)
... fig.colorbar(mpl.cm.ScalarMappable(cmap=cmap3, norm=norm3),
... cax=axl3,
... # to use 'extend', you must specify two extra boundaries:
... boundaries=[0] + bounds + [18],
... extend='both',
... ticks=bounds, # optional
... spacing='proportional',
... orientation='horizontal',
... label='Discrete intervals, some other units')
...
...
... # Create the 17 segment model
... bullseye_plot(ax[0], data, cmap=cmap, norm=norm)
... ax[0].set_title('Bulls Eye (AHA)')
...
... bullseye_plot(ax[1], data, cmap=cmap2, norm=norm2)
... ax[1].set_title('Bulls Eye (AHA)')
...
... bullseye_plot(ax[2], data, seg_bold=[3, 5, 6, 11, 12, 16],
... cmap=cmap3, norm=norm3)
... ax[2].set_title('Segments [3, 5, 6, 11, 12, 16] in bold')
...
... plt.show()
...