>>> """
=================
Contourf Hatching
=================
Demo filled contour plots with hatched patterns.
"""
... import matplotlib.pyplot as plt
... import numpy as np
...
... # invent some numbers, turning the x and y arrays into simple
... # 2d arrays, which make combining them together easier.
... x = np.linspace(-3, 5, 150).reshape(1, -1)
... y = np.linspace(-3, 5, 120).reshape(-1, 1)
... z = np.cos(x) + np.sin(y)
...
... # we no longer need x and y to be 2 dimensional, so flatten them.
... x, y = x.flatten(), y.flatten()
...
... ###############################################################################
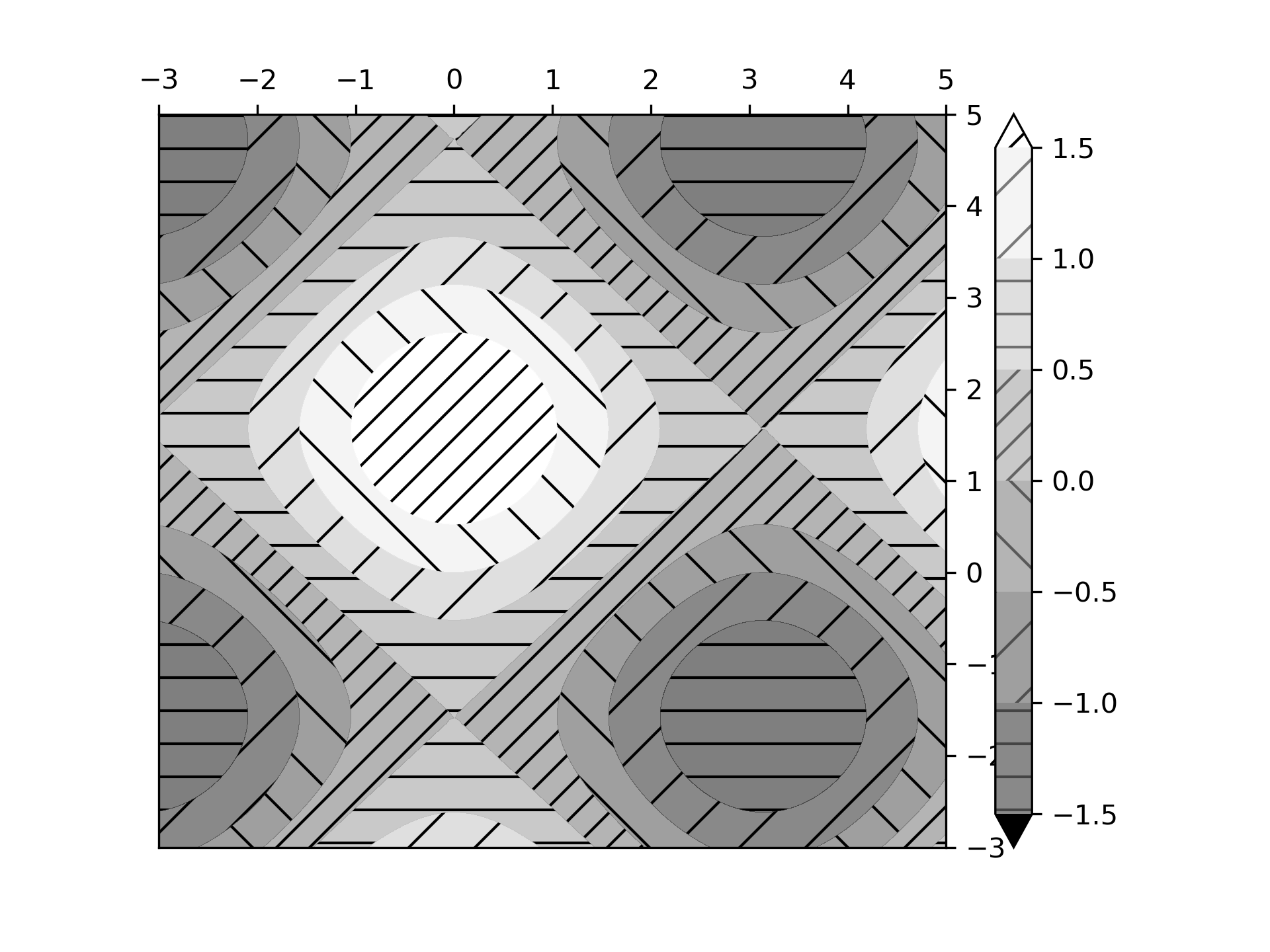
... # Plot 1: the simplest hatched plot with a colorbar
...
... fig1, ax1 = plt.subplots()
... cs = ax1.contourf(x, y, z, hatches=['-', '/', '\\', '//'],
... cmap='gray', extend='both', alpha=0.5)
... fig1.colorbar(cs)
...
... ###############################################################################
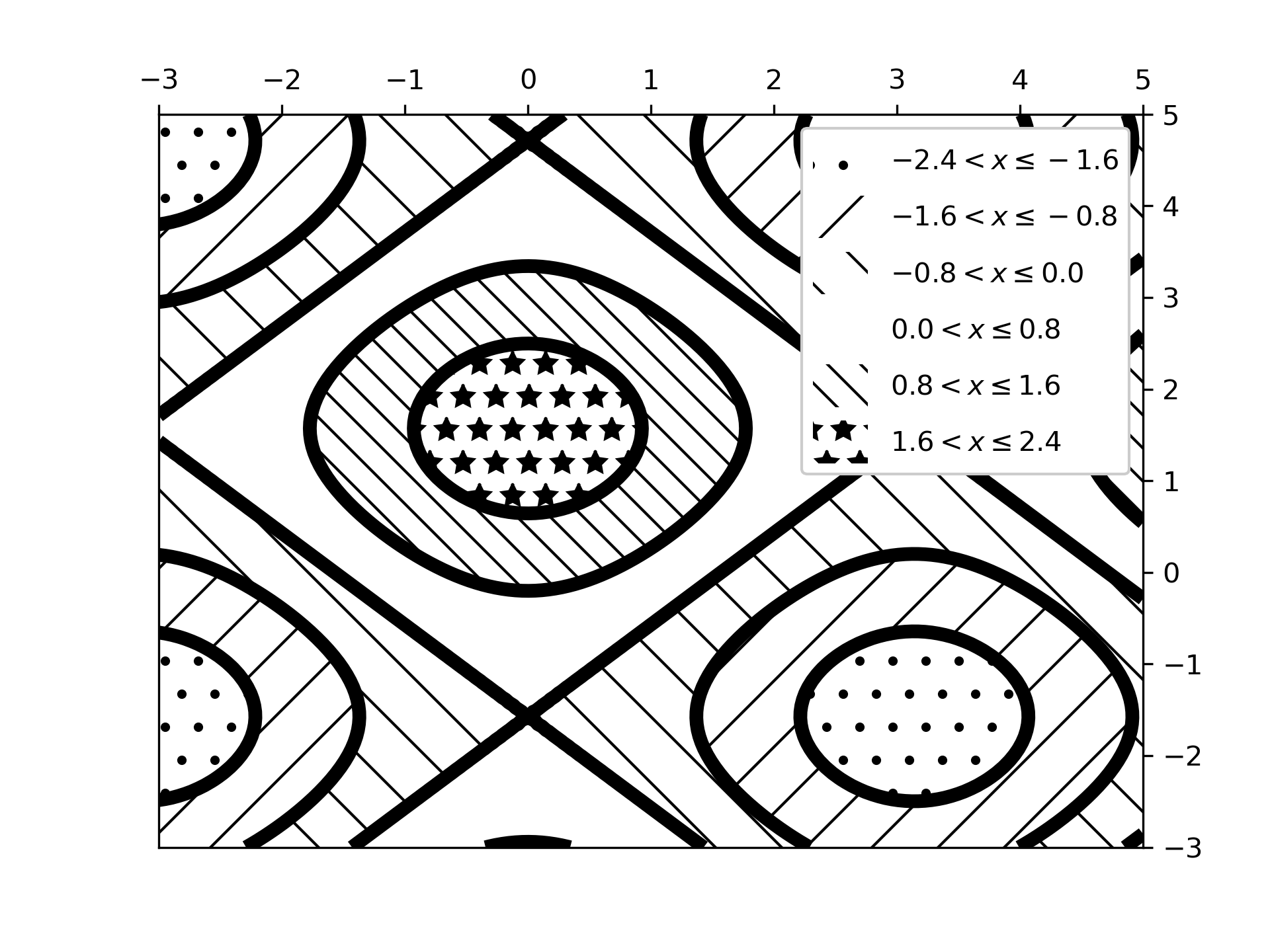
... # Plot 2: a plot of hatches without color with a legend
...
... fig2, ax2 = plt.subplots()
... n_levels = 6
... ax2.contour(x, y, z, n_levels, colors='black', linestyles='-')
... cs = ax2.contourf(x, y, z, n_levels, colors='none',
... hatches=['.', '/', '\\', None, '\\\\', '*'],
... extend='lower')
...
... # create a legend for the contour set
... artists, labels = cs.legend_elements(str_format='{:2.1f}'.format)
... ax2.legend(artists, labels, handleheight=2, framealpha=1)
... plt.show()
...
... #############################################################################
... #
... # .. admonition:: References
... #
... # The use of the following functions, methods, classes and modules is shown
... # in this example:
... #
... # - `matplotlib.axes.Axes.contour` / `matplotlib.pyplot.contour`
... # - `matplotlib.axes.Axes.contourf` / `matplotlib.pyplot.contourf`
... # - `matplotlib.figure.Figure.colorbar` / `matplotlib.pyplot.colorbar`
... # - `matplotlib.axes.Axes.legend` / `matplotlib.pyplot.legend`
... # - `matplotlib.contour.ContourSet`
... # - `matplotlib.contour.ContourSet.legend_elements`
...